Recently, I spent some time on exploring the feature of SAS ODS Graphics. ODS Graphics offers an easy way to generate high-quality statistical graphics without extensive programming using SAS/Graph. ODS Graphics has been included in almost all of SAS/Stat Procedure.
In the discussions below, the example from SAS Proc Lifetest is used to illustrate the generation of Kaplan-Meier curve using ODS Graphics.
On the basis of SAS Example 49.2 Enhanced Survival Plot and Multiple-Comparison Adjustments, we can run the following program.
proc format;
value risk 1='ALL' 2='AML-Low Risk' 3='AML-High Risk';
data BMT;
input Group T Status @@;
format Group risk.;
label T='Disease Free Time';
datalines;
1 2081 0 1 1602 0 1 1496 0 1 1462 0 1 1433 0
1 1377 0 1 1330 0 1 996 0 1 226 0 1 1199 0
1 1111 0 1 530 0 1 1182 0 1 1167 0 1 418 1
1 383 1 1 276 1 1 104 1 1 609 1 1 172 1
1 487 1 1 662 1 1 194 1 1 230 1 1 526 1
1 122 1 1 129 1 1 74 1 1 122 1 1 86 1
1 466 1 1 192 1 1 109 1 1 55 1 1 1 1
1 107 1 1 110 1 1 332 1 2 2569 0 2 2506 0
2 2409 0 2 2218 0 2 1857 0 2 1829 0 2 1562 0
2 1470 0 2 1363 0 2 1030 0 2 860 0 2 1258 0
2 2246 0 2 1870 0 2 1799 0 2 1709 0 2 1674 0
2 1568 0 2 1527 0 2 1324 0 2 957 0 2 932 0
2 847 0 2 848 0 2 1850 0 2 1843 0 2 1535 0
2 1447 0 2 1384 0 2 414 1 2 2204 1 2 1063 1
2 481 1 2 105 1 2 641 1 2 390 1 2 288 1
2 421 1 2 79 1 2 748 1 2 486 1 2 48 1
2 272 1 2 1074 1 2 381 1 2 10 1 2 53 1
2 80 1 2 35 1 2 248 1 2 704 1 2 211 1
2 219 1 2 606 1 3 2640 0 3 2430 0 3 2252 0
3 2140 0 3 2133 0 3 1238 0 3 1631 0 3 2024 0
3 1345 0 3 1136 0 3 845 0 3 422 1 3 162 1
3 84 1 3 100 1 3 2 1 3 47 1 3 242 1
3 456 1 3 268 1 3 318 1 3 32 1 3 467 1
3 47 1 3 390 1 3 183 1 3 105 1 3 115 1
3 164 1 3 93 1 3 120 1 3 80 1 3 677 1
3 64 1 3 168 1 3 74 1 3 16 1 3 157 1
3 625 1 3 48 1 3 273 1 3 63 1 3 76 1
3 113 1 3 363 1
;
ods rtf file="c:\temp\test.doc" style=journal;
ods graphics on;
ods trace on;
proc lifetest data=BMT plots=survival(atrisk=0 to 2500 by 500);
ods select SurvivalPlot;
time T * Status(0);
strata Group / test=logrank adjust=sidak;
run;
ods trace off;
ods graphics off;
ods rtf close;
In the above statements,
ods rtf file="c:\temp\test.doc" specifies the ODS output as RTF file and the file location and file name. style=journal specifies the use of output in publication quality.
"ods graphics on" to invoke the ODS Graphics
"ods trace on" is not really necessary, but it is useful when we need to know the name/location of the SAS template. If we check the SAS Log Window, we will be able to see that the SAS template for the corresponding graph is Stat.Lifetest.Graphics.ProductLimitSurvival
Output Added:
-------------
Name: SurvivalPlot
Label: Survival Curves
Template: Stat.Lifetest.Graphics.ProductLimitSurvival
Path: Lifetest.SurvivalPlot
-------------
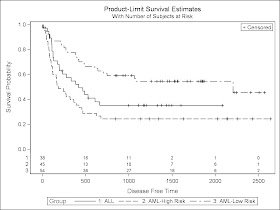
PROC LIFETEST is invoked to compute the product-limit estimate of the survivor function for each of three risk categories (1='ALL' 2='AML-Low Risk' 3='AML-High Risk'). Using ODS Graphics, you can display the number of subjects at risk in the survival plot. The PLOTS= option requests that the survival curves be plotted, and the ATRISK= suboption specifies the time points at which the at-risk numbers are displayed. In the STRATA statement, the ADJUST=SIDAK option requests the
After the run, we will need to use the following statements to close ODS.
ods trace off;
ods graphics off;
ods rtf close;
The above program will create a high quality figure like below:
Suppose we would like to change the title and labels of the figure above, we can use PROC Template. First thing we need to do is to identify which Template we need to modify. The SAS Log window from ods trace on statement indicates the following template.
Stat.Lifetest.Graphics.ProductLimitSurvival
To open this template, we would need to the pull down manu in SAS window, then choose View -> Results -> View -> Template
From Template window, we will locate the following folder:
SAShelp.tmplmst -> stat -> lifetest -> graphics
We will then see the template called ProductLimitSurvival. Double click this file, we will be able to see the contents in this template file.
The template file can be copied and pasted into the program window. We can then edit the template file to modify the title, labels, legends,…
Suppose we run the following program to generate the Kaplan-Meier curve (upward curve instead of the original downward curve). The upward curve is sometimes easier to be understood. For example, it is useful in the time to event variable where the event is a better outcome (for example, time to recovery, time to release from the hospital, time to immune tolerance (in Hemophilia). By changing the layout of the figure, we would also need to modify the label.
proc lifetest data=BMT plots=survival(f test);
ods select failurePlot;
time T * Status(0);
strata Group / test=logrank adjust=sidak;
run;
plots=survival(f test) requests the failure Plot (upward) instead of the survival plot (downward). The ‘test’ indicates the display of the logrank test results.
From the LOG window trigged by ODS Trace On statement, we will be able to know that the template is Stat.Lifetest.Graphics.ProductLimitFailure which is located in the same folder as Stat.Lifetest.Graphics.ProductLimitSurvival discussed before.
Output Added:
-------------
Name: FailurePlot
Label: Failure Curves
Template: Stat.Lifetest.Graphics.ProductLimitFailure
Path: Lifetest.FailurePlot
We can copy and paste the template statements and run as part of the program.
ods rtf file="c:\temp\test.doc" style=journal;
ods graphics on;
ods trace on;
proc template;
define statgraph Stat.Lifetest.Graphics.ProductLimitFailure;
dynamic NStrata xName maxTime plotAtRisk plotCensored plotCL plotHW
plotEP labelCL labelHW labelEP yMin StratumID classAtRisk plotTest
GroupName Transparency SecondTitle TestName pValue;
BeginGraph;
if (NSTRATA=1)
if (EXISTS(STRATUMID))
entrytitle "Kaplan-Meier Curve" " for " STRATUMID; /*Revised the Figure title*/
else
entrytitle "Kaplan-Meier Curve";
endif;
if (PLOTATRISK)
entrytitle "with Number of Subjects at Risk" / textattrs=
GRAPHVALUETEXT;
endif;
layout overlay / xaxisopts=(label="Time to Recovery" offsetmin=.05 linearopts /*xaxisopts=(label=" ") to change the x-axis label*/
=(viewmax=MAXTIME)) yaxisopts=(label="Probability of Achieving Tolerance"
shortlabel="Failure" linearopts=(viewmin=0 viewmax=1 tickvaluelist
=(0 .2 .4 .6 .8 1.0)));
if (PLOTHW=1 AND PLOTEP=0)
bandplot LimitUpper=eval (1-HW_LCL) LimitLower=eval (1-HW_UCL)
x=TIME / modelname="Failure" fillattrs=GRAPHCONFIDENCE name=
"HW" legendlabel=LABELHW;
endif;
if (PLOTHW=0 AND PLOTEP=1)
bandplot LimitUpper=eval (1-EP_LCL) LimitLower=eval (1-EP_UCL)
x=TIME / modelname="Failure" fillattrs=GRAPHCONFIDENCE name=
"EP" legendlabel=LABELEP;
endif;
if (PLOTHW=1 AND PLOTEP=1)
bandplot LimitUpper=eval (1-HW_LCL) LimitLower=eval (1-HW_UCL)
x=TIME / modelname="Failure" fillattrs=GRAPHDATA1
datatransparency=.55 name="HW" legendlabel=LABELHW;
bandplot LimitUpper=eval (1-EP_LCL) LimitLower=eval (1-EP_UCL) x=
TIME / modelname="Failure" fillattrs=GRAPHDATA2
datatransparency=.55 name="EP" legendlabel=LABELEP;
endif;
if (PLOTCL=1)
if (PLOTHW=1 OR PLOTEP=1)
bandplot LimitUpper=eval (1-SDF_LCL) LimitLower=eval (1-SDF_UCL
) x=TIME / modelname="Failure" display=(outline) outlineattrs=
GRAPHPREDICTIONLIMITS name="CL" legendlabel=LABELCL;
else
bandplot LimitUpper=eval (1-SDF_LCL) LimitLower=eval (1-SDF_UCL
) x=TIME / modelname="Failure" fillattrs=GRAPHCONFIDENCE name=
"CL" legendlabel=LABELCL;
endif;
endif;
stepplot y=eval (1-SURVIVAL) x=TIME / name="Failure" rolename=(
_tip1=ATRISK _tip2=EVENT) tip=(y x Time _tip1 _tip2)
legendlabel="Failure";
if (PLOTCENSORED)
scatterplot y=eval (1-CENSORED) x=TIME / markerattrs=(symbol=
plus) name="Censored" legendlabel="Censored";
endif;
if (PLOTCL=1 OR PLOTHW=1 OR PLOTEP=1)
discretelegend "Censored" "CL" "HW" "EP" / location=outside
halign=center;
else
if (PLOTCENSORED=1)
discretelegend "Censored" / location=inside autoalign=(topleft
bottomright);
endif;
endif;
if (PLOTATRISK=1)
innermargin / align=bottom;
blockplot x=TATRISK block=ATRISK / repeatedvalues=true display=
(values) valuehalign=start valuefitpolicy=truncate
labelposition=left labelattrs=GRAPHVALUETEXT valueattrs=
GRAPHDATATEXT (size=7pt) includemissingclass=false;
endinnermargin;
endif;
endlayout;
else
entrytitle "Kaplan-Meier Curve";
if (EXISTS(SECONDTITLE))
entrytitle SECONDTITLE / textattrs=GRAPHVALUETEXT;
endif;
layout overlay / xaxisopts=(label="Time to Recovery" offsetmin=.05 linearopts
=(viewmax=MAXTIME)) yaxisopts=(label="Probability of Recovery" /*y-axis label*/
shortlabel="Failure" linearopts=(viewmin=0 viewmax=1 tickvaluelist
=(0 .2 .4 .6 .8 1.0)));
if (PLOTHW=1)
bandplot LimitUpper=eval (1-HW_LCL) LimitLower=eval (1-HW_UCL)
x=TIME / group=STRATUM index=STRATUMNUM modelname="Failure"
datatransparency=Transparency;
endif;
if (PLOTEP=1)
bandplot LimitUpper=eval (1-EP_LCL) LimitLower=eval (1-EP_UCL)
x=TIME / group=STRATUM index=STRATUMNUM modelname="Failure"
datatransparency=Transparency;
endif;
if (PLOTCL=1)
if (PLOTHW=1 OR PLOTEP=1)
bandplot LimitUpper=eval (1-SDF_LCL) LimitLower=eval (1-SDF_UCL
) x=TIME / group=STRATUM index=STRATUMNUM modelname="Failure"
display=(outline);
else
bandplot LimitUpper=eval (1-SDF_UCL) LimitLower=eval (1-SDF_LCL
) x=TIME / group=STRATUM index=STRATUMNUM modelname="Failure"
datatransparency=Transparency;
endif;
endif;
stepplot y=eval (1-SURVIVAL) x=TIME / group=STRATUM index=
STRATUMNUM name="Failure" rolename=(_tip1=ATRISK _tip2=EVENT)
tip=(y x Time _tip1 _tip2);
if (PLOTCENSORED)
scatterplot y=eval (1-CENSORED) x=TIME / group=STRATUM index=
STRATUMNUM markerattrs=(symbol=plus);
endif;
if (PLOTATRISK)
innermargin / align=bottom;
blockplot x=TATRISK block=ATRISK / class=CLASSATRISK
repeatedvalues=true display=(label values) valuehalign=start
valuefitpolicy=truncate labelposition=left labelattrs=
GRAPHVALUETEXT valueattrs=GRAPHDATATEXT (size=7pt)
includemissingclass=false;
endinnermargin;
endif;
DiscreteLegend "Failure" / title="Risk Group" location=inside autoalign=(bottomright); /*Revise the legend*/
if (PLOTCENSORED)
if (PLOTTEST)
layout gridded / rows=2 autoalign=(TOPLEFT BOTTOMRIGHT BOTTOM
TOP) border=true BackgroundColor=GraphWalls:Color Opaque=true;
entry "+ Censored";
if (PVALUE < .0001)
entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
else
entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
endif;
endlayout;
else
layout gridded / rows=1 autoalign=(TOPLEFT BOTTOMRIGHT BOTTOM
TOP) border=true BackgroundColor=GraphWalls:Color Opaque=true;
entry "+ Censored";
endlayout;
endif;
else
if (PLOTTEST)
layout gridded / rows=1 autoalign=(TOPLEFT BOTTOMRIGHT BOTTOM
TOP) border=true BackgroundColor=GraphWalls:Color Opaque=true;
if (PVALUE < .0001)
entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
else
entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
endif;
endlayout;
endif;
endif;
endlayout;
endif;
EndGraph;
end;
run;
proc lifetest data=BMT plots=survival(f test);
ods select quartiles FailurePlot;
time T * Status(0);
strata Group / test=logrank adjust=sidak;
run;
ods trace off;
ods graphics off;
ods rtf close;
At the end of the program, we use the following program to delete the user defined Template.
proc template;
delete Stat.Lifetest.Graphics.ProductLimitFailure;
run;
Further reading:
- Getting Started with ODS Statistical Graphics in SAS® 9.2 presents the fundamental information you need to get started with ODS Statistical Graphics in SAS 9.2. (Revised 2009)
- Summary of ODS Graphics Capabilities provides a concise overview of the capabilities of ODS Graphics.


Does anyone know how to create a heaviside function in SAS when the predictor variable has 3 categories?
ReplyDeleteIn all clinical studies, collecting reliable adverse event data is critical. It is quite usual to gather unfavorable occurrences and their features (including seriousness, severity, causality to the study drug, and so on).
ReplyDelete